One of the most fundamental processes in cells, which ensures genomic stability, is accurate DNA synthesis. Error-free duplication of the human genome is crucial to prevent genomic instability. Incomplete or incorrect DNA replication induces breaks and mutations in DNA, which could transform a normal cell into a cancer cell or induce diseases, such as repeat expansion disorders. Our goal is to dissect the DNA replication in human cells at the endogenous locus in vivo to identify replication defects, which could lead to genomic instability and diseases.
DNA Replication Defects in Human Cells
To dissect the DNA replication program at the endogenous disease locus and find molecular components leading to genomic instability and DNA mutations in human cells we are using a unique technique, called single-molecule analysis of replicated DNA (SMARD). SMARD allows us to visualize the DNA replication at the endogenous genomic locus in human cells at single DNA molecule level. Using SMARD we revealed an altered replication program in FXS human embryonic stem cells (hESCs) and in FRDA induced pluripotent stem cells (iPSCs). In addition we found that stalled replication forks are driving GAA repeat expansion in FRDA cells. It is important to understand which defects in the replication occur in the native chromosomal context for the development of effective therapeutic treatments to prevent mutations in patients with diseases like FRDA and FXS.
We are in particular interested in stem cells. We have a collection of embryonic and induced pluripotent stem cells derived from several diseases, such as FXS as well as from carriers, containing a BRCA1 or BRCA2 germline mutation.
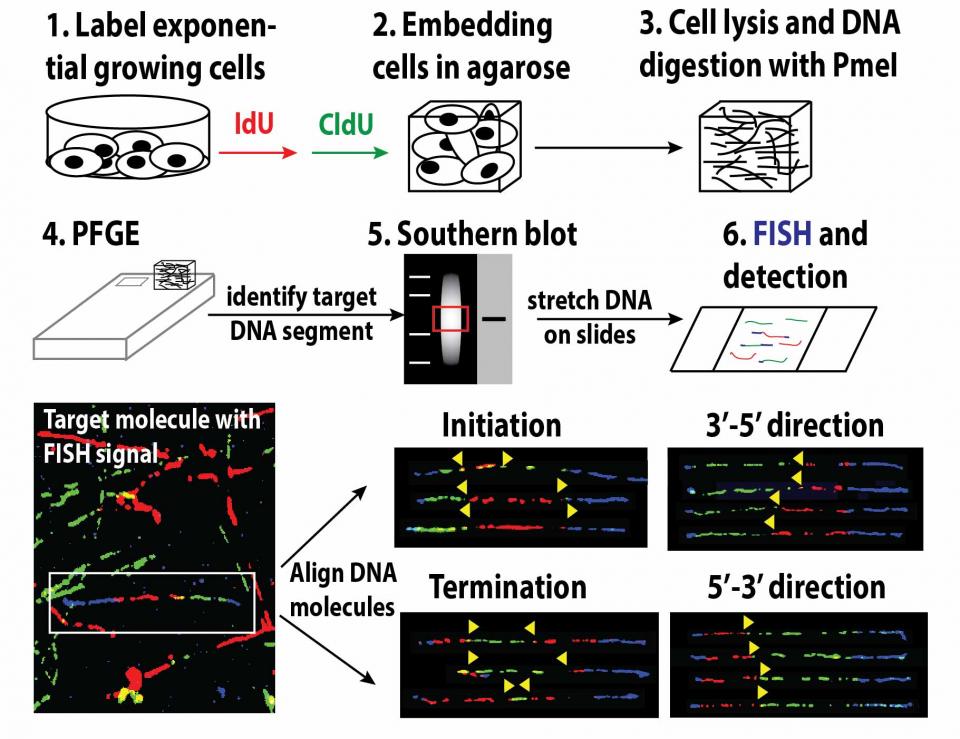
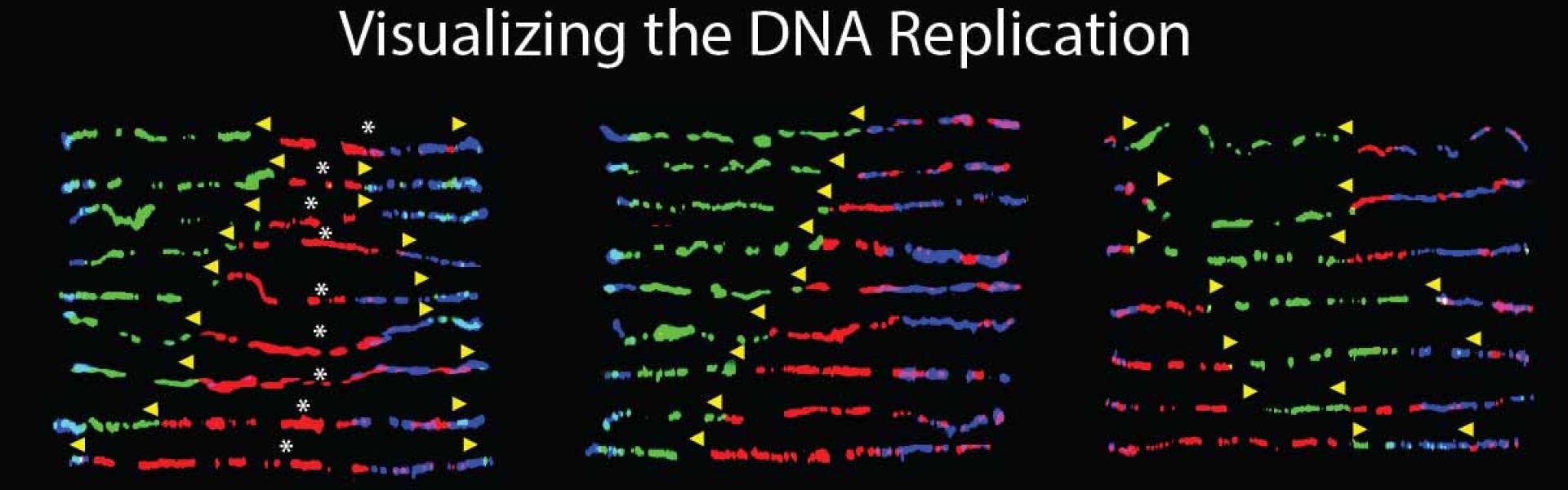
SMARD, single-molecule analysis of replicated DNA
Exponentially growing cells are sequentially pulsed with two different halogenated nucleosides to label replicating DNA. Pulsed cells are embedded in agarose plugs, lysed, and genomic DNA is digested with a rare cutting restriction endonuclease to produce segments of 100 – 600 kb. The digested DNA is then separated according to size by pulsed-field gel electrophoresis and the target segment within the gel is identified by Southern blot. A gel slice containing the target segment is excised from the gel and melted. The DNA is then stretched on silanized glass slides and the halogenated nucleosides incorporated in the replicated DNA are detected by immunostaining. Biotinylated FISH probes are used to identify the target molecules and to align the images of individual molecules to produce a composite profile of replication.