DNA Replication Defects in Cancer and Stem Cells
The mechanism(s) facilitating mutagenesis and causing genomic instability in human cells are not clearly understood. Accurate, error-free duplication of the human genome is crucial to prevent mutations, which could lead to carcinogenesis. To study the early steps triggering mutagenesis we are using human embryonic stem cells (hESCs) derived from embryos with a germline mutation known to induce cancer, such as BRCA1 and BRCA2. Breast cancer is the leading cause of cancer in women in the United States and the third leading cause of cancer death. Germline mutations in one copy of the BRCA1 or BRCA2 genes account for the majority of hereditary breast cancers and are found in approximately 1 in 250 individuals in the general population. These mutations are associated with highly elevated lifetime risks of breast cancer (approximately 70-80%) and ovarian cancer (45% for BRCA1 and 20% for BRCA2 carriers). Risk-reducing surgery is the most effective way to avoid breast cancer, however only a minority of women choose prophylactic mastectomy as prevention. So far, there are not enough mechanistic insights to formulate new preventive drugs for BRCA1 and BRCA2 carriers. Knowledge about the early mechanisms causing genomic instability in BRCA carriers would enable us to derive strategies to impede mutagenesis in these patients. Our goal is to identify the early steps that cause genomic instability and mutagenesis in BRCA1 and 2 carriers in order to find preventive strategies.
Fragile X syndrome and Fragile X-Associated Diminished Ovarian Reserve
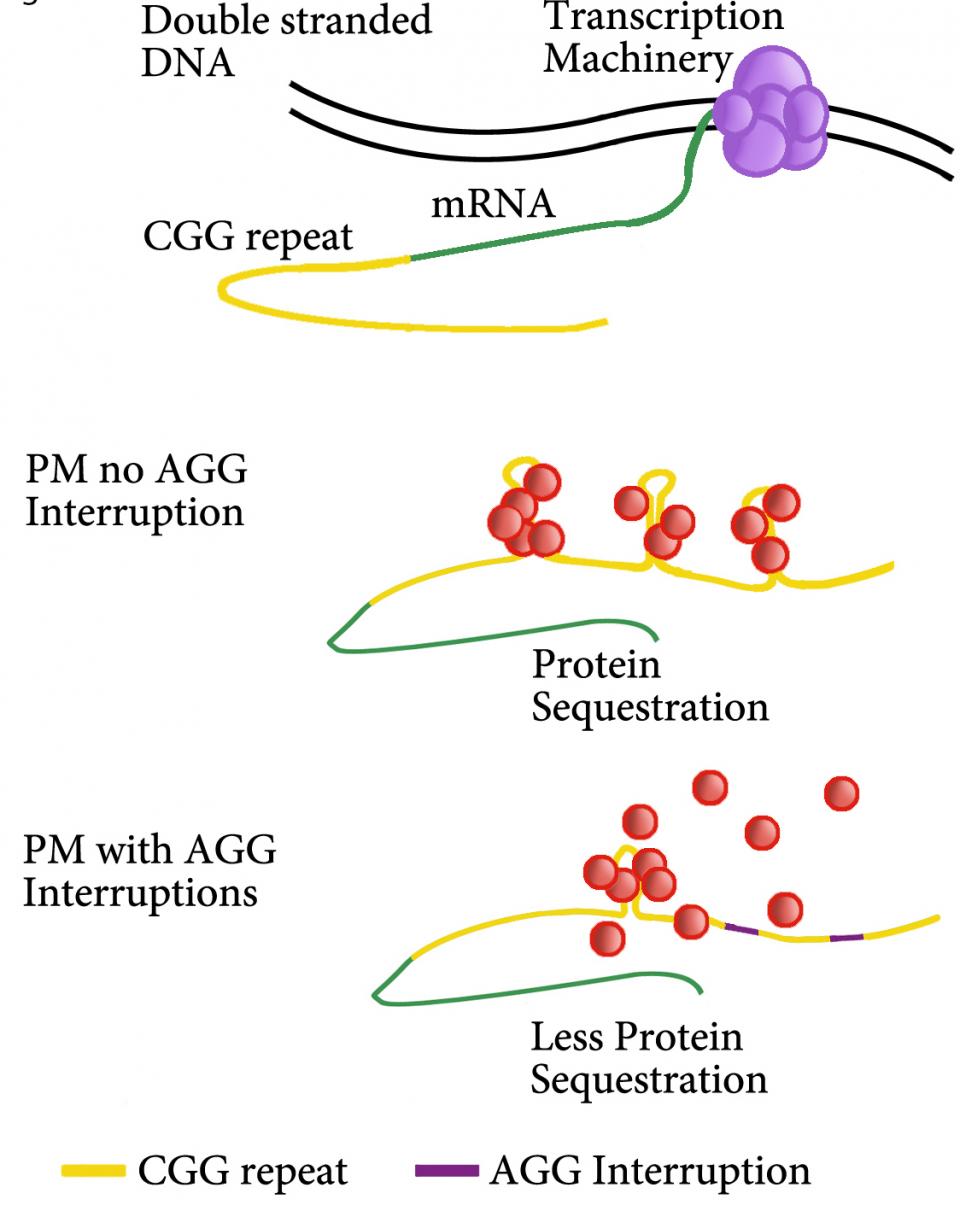
Fragile X syndrome (FXS) (OMIM #300624) is the most common cause of heritable intellectual disability and is the leading single-gene defect associated with autism. FXS is caused by a CGG repeat expansion in the 5′UTR of the fragile X mental retardation (FMR1) gene on the X chromosome. However, besides FXS, expansion of CGG repeats within the FMR1 gene is also associated with other disorders: fragile X–associated primary ovarian insufficiency (FXPOI) and fragile X–associated diminished ovarian reserve (FXDOR). Whereas FXS is caused by CGG repeat length of over 200 (full mutation (FM)) and FMR1 gene silencing, it has been proposed that FXPOI and FXDOR are caused by toxic FMR1 messenger RNA (mRNA) and/or expression of an aberrant FMRpolyG protein in premutation (PM) patients with 55–200 repeats. We found that carriers with mid-range repeats size (70-90 CGG) demonstrate significantly lower ovarian reserve. Additionally, patients with less AGG interruptions had the lowest ovarian reserve. Uninterrupted CGG repeats (yellow) could form secondary structures within ovarian cells such as granulosa cells. These RNA structures could cause binding and sequestering of RNA-binding proteins (red balls), which function would be lost in the cells causing cell toxicity. AGG interruptions (purple) in the repeat sequence could prevent the formation of these secondary repeat structures, thus lower the sequestering of proteins in the cell. This in turn could results in less accumulation of proteins in the cells, and could prevent cell dysfunction in the ovary in women carrying a PM.
DNA Replication Defects in Repeat Expansion Disorders
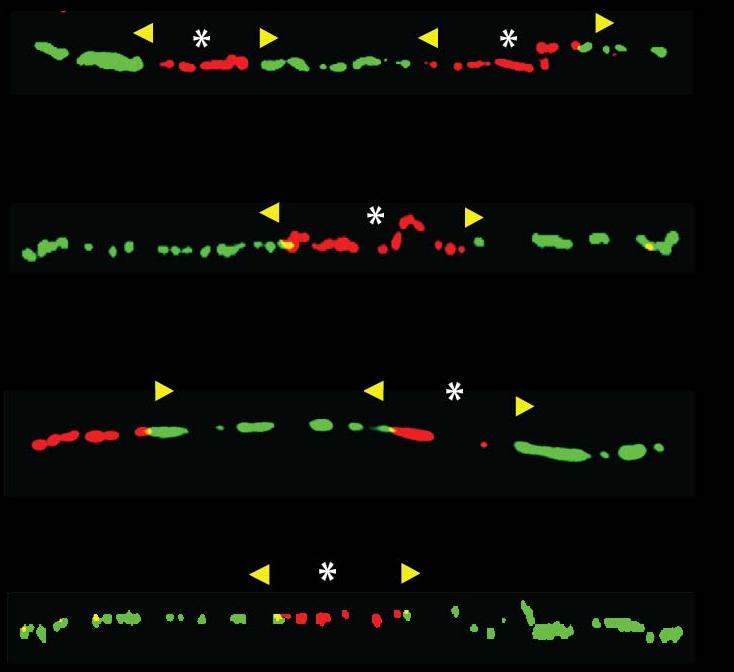
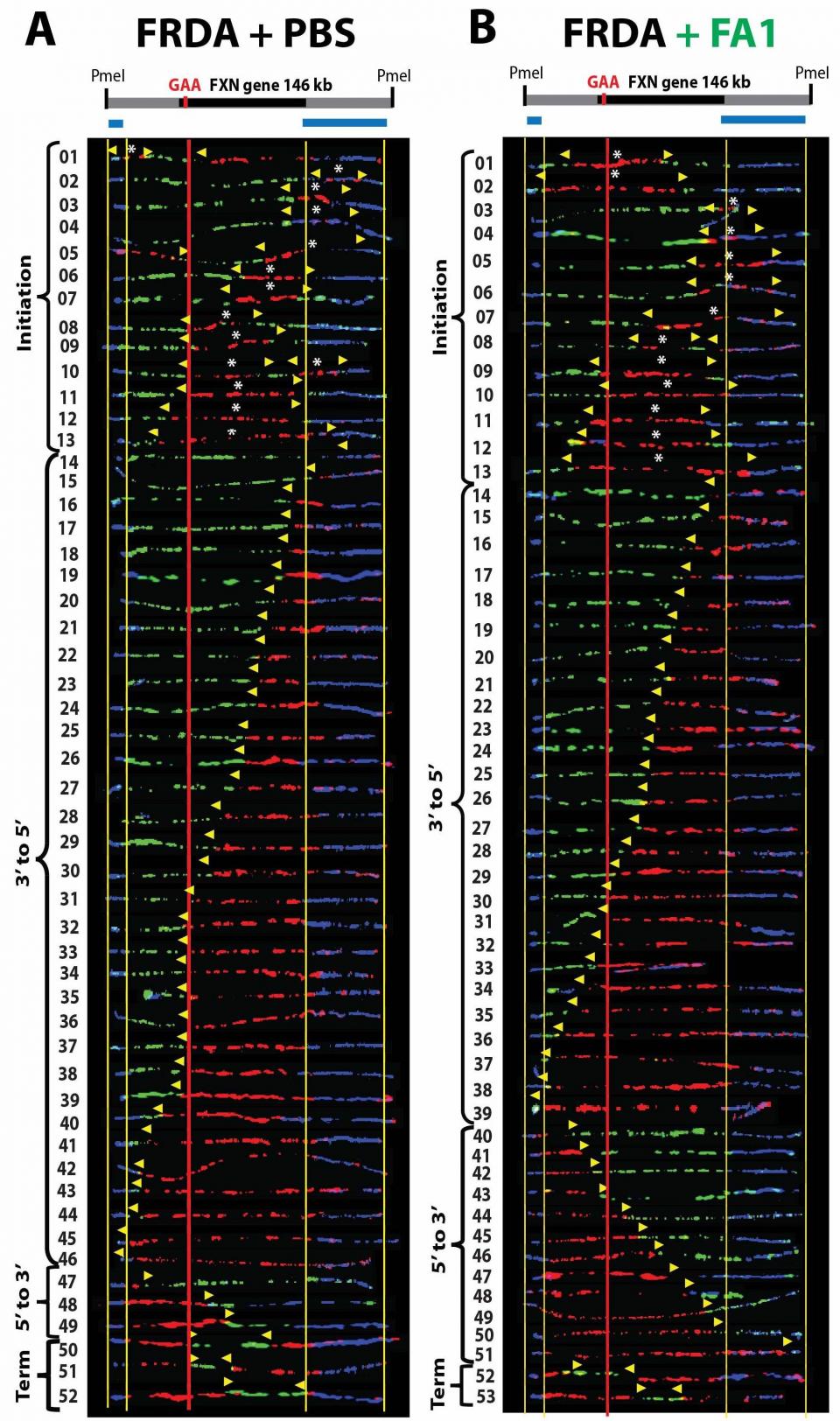
Several inherited human neurological and muscular disorders like Friedreich’s ataxia (FRDA), fragile X syndrome (FXS), myotonic dystrophy (DM1), Huntington’s disease and spinocerebellar ataxia type 7 (SCA7) are caused by a trinucleotide repeat expansion. The mechanism leading to repeat instability is still not known and there is currently no cure for these repeat expansion diseases. One goal of our research is to dissect the DNA replication program at the endogenous disease locus and find molecular components driving repeat expansion in human cells. Therefore we are using a unique technique, called single-molecule analysis of replicated DNA (SMARD), which allows us to visualize the DNA replication at the endogenous genomic locus in human cells at single DNA molecule level. Using SMARD we revealed an altered replication program in FXS human embryonic stem cells (hESCs) and in FRDA induced pluripotent stem cells (iPSCs). We were able, for the first time, to visualize errors in the DNA replication at the endogenous FMR1 and FXN gene locus. We found altered replication fork progression and stalled replication forks at the repeats in FXS and FRDA cells. It is important to understand how repeat expansion occurs in the native chromosomal context for the development of effective therapeutic treatments to prevent repeat expansions in patients with diseases like FRDA and FXS. For example, we detected a major replication fork stall at the expanded GAA repeats in Friedreich’s ataxia iPSCs, which we were able to release with small molecules that also stabilize the GAA repeats to prevent repeat expansion.
https://www.ncbi.nlm.nih.gov/pubmed/24289922
https://www.ncbi.nlm.nih.gov/pubmed/?term=gerhardt+napierala